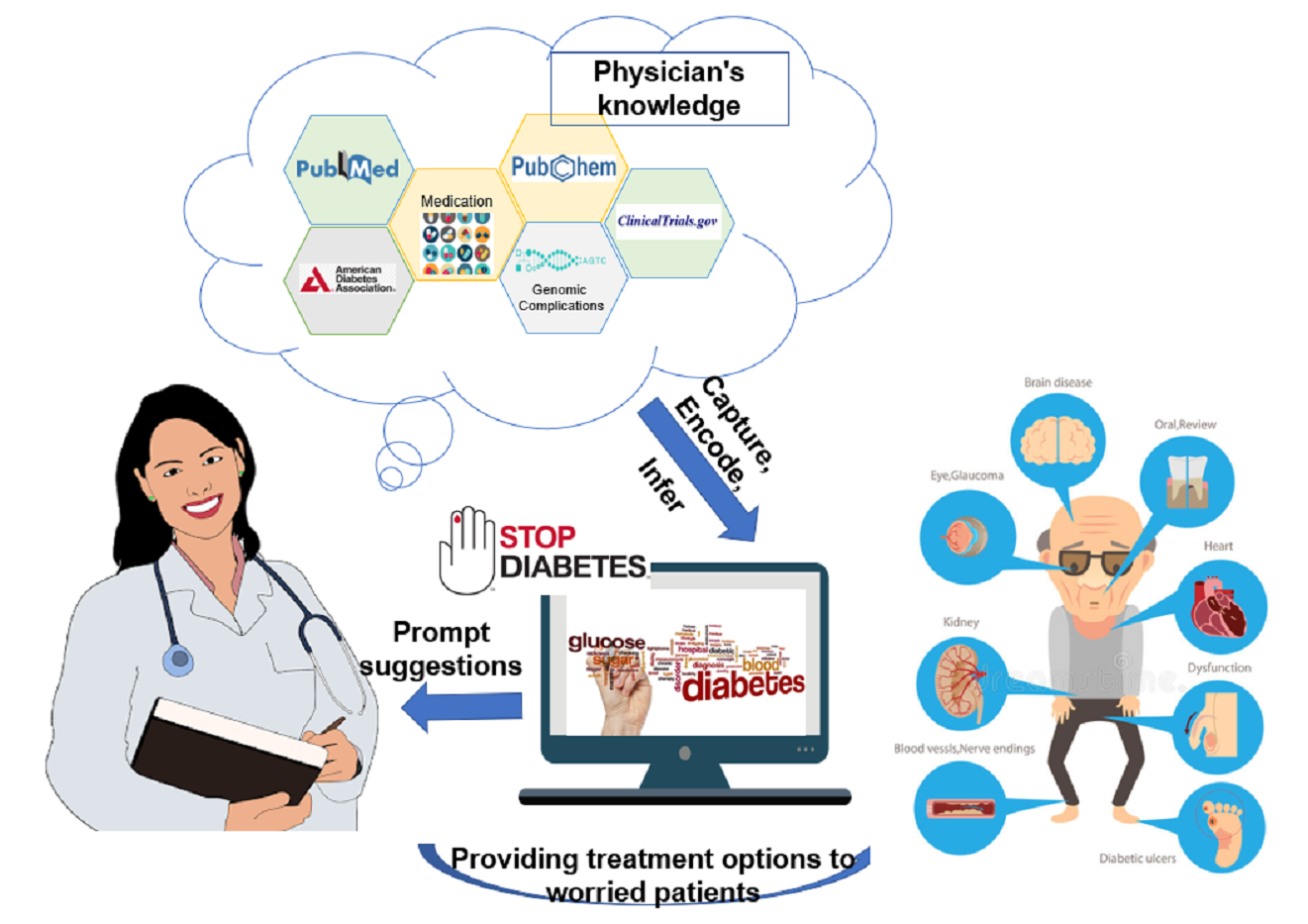
The number of people living with diabetes mellitus is constantly on the rise, from 108 million in 1980 to 422 million in 2014. And it’ll be the seventh leading cause of death in 2030. The need to rapidly and effectively diagnose and treat the illness is crucial. Recommendations in authoritative literature, Clinical Practice Guidelines (CPG)’s are often tailored to a diverse populations, but as patients get more complex they might not align well with supporting evidence. As a result, one of the challenges that physicians face in using CPG recommendations is determining how well the study evidence applies to a general clinical population. We want to assist the physician in identifying gaps within guidelines to make a informed tailored treatment recommendation. Once a patient alignment has been ascertained, we want to help the physician group the patient into different therapies (initial, dual and triple therapy), taking into consideration the patient comorbidities, previous patient history and drug complications. We aim for our system to make the physician aware of these adverse effects of clinically tested drugs - particularly to cardiotoxicity, and also we want to account for drug conflicts with comorbid conditions. Finally, the system will assign provenance (evidence) to the discovered treatment recommendations for the patient, and will be able to monitor the patient’s condition (both prescribed medicine and the prescribed diet) over time
Significance
General
- Integrate heterogeneous knowledge sources
- Assistance in decision making for complicated patients
Parts
Drug recommendation: With a lot of information available on antidiabetic drugs, and a very comprehensive and detailed ADA guidelines to follow, it is hard for a physician that has little time and a wide range of patients, to find a suggested drug tailored to a patient. This system makes this information easy to access.
Toxicity: There is a lot of toxicity information for drugs available on many different sources, Pubchem, DrugBank, ToxBank, however for a user that is not aware of all of the resources or has the time to go through all of the resources, having all of this information combined by semantics, lets the user easily access this information.
Cohort: It is important to know whether a patient that we are suggesting a drug to is represented in the studies that we are getting this suggestion from. It provides provenance and trust to the user that a suggested drug would suit a particular patient.
Integration of the above parts: having all of this information connected and easy to use is very helpful. A physician not only needs to know a suggested drug for a particular patient profile, they also need to know possible side effects of it, and whether the patient they are recommending to is well represented in the studies they are using. All of this information combined in one ontology, makes our ontology very powerful and useful. Combining the parts together, there are also various portions that are interconnected. For instance, toxicity of a drug can also be based on particular clinical trials on humans - our system can connect this to a cohort and compare our patient to this cohort as well.
Developers
- Bhanushee Sharma : sharmb3 at rpi dot edu
- Neha Keshan : keshan at rpi dot edu
- Nkechinyere Nneka Agu : agun at rpi dot edu
- Shruthi Chari : charis at rpi dot edu
- Shweta Narkar : narkas at rpi dot edu
We would be delighted to have people use our ontologies, review the results and contribute to our work. Several components of this project reflect ongoing research, and although the course has ended we expect the work to be reused and extended over the next several years. If you are interested in getting involved, please review our Getting Involved and Maintenance Policy below and feel free to reach out to us with any feedback or to let us know you would like to work with us on any aspect of the project.
Acknowledgements
We would like to thank Prof. Deborah McGuiness, Ms Elisa Kendall, Dr Jim McCusker for their continuing support throughout the course of this project. We would also like to thank Dr Oshani Seneviratne for her inputs and Dr Amar Das from IBM Research, Prof. Jonathan S. Dordick and Dr. Keith Fraser of Center for Biotechnology and Interdisciplinary Studies at Rensselaer Polytechnic Institute.
This was developed as a part of the Ontology Engineering course supervised by Prof. Deborah McGuinness and Ms. Elisa Kendall at RPI in Fall'18
How you can contribute to Diabetes Treatment Support Ontology
Our ontology at present is successful at taking in a patient’s parameters, selecting a drug for them, displaying the drug’s toxicity, selecting an alternative drug for the initial drug, displaying its toxicity and determining whether that particular patient group or cohort has been studied in the ADA guidelines. Going forward, there are various ways we can get the broader community involved in our project.
Toxicity
The current system we have in place for toxicity is quite limited, we are only looking at two drug’s toxicity (Metformin and Phenformin), and manually extracting this information from PubChem snippets. You could help us in automating this process, and not only looking at Pubchem, but a variety of papers in PubMed. We also need help with finding all the relevant toxicity information on a drug and its similar drug - in deciding which studies and toxicities would be useful and relevant. There are various other portions we could use help in expanding our system - for instance in adding metabolite information to the drugs and their possible toxicities or adding contraindication scenarios to the system (for instance for a patient that would have a contraindication to Metformin). We could also use the help of domain experts from the medical field in order to give guidance on which toxicity information would be directly useful to them. However, if we were to expand this system, understanding the question of connecting drugs to their metabolites and their toxicities could be more broadly useful in the field of toxicology, as it can be applied to many other diseases and drugs - and perhaps could find links in certain drug structures and their toxicities from metabolites, which could be helpful to be aware of when designing new drugs.
Recommendation
As of now, the recommendation is really simple and only focuses on the pharmaceutical section of ADA guideline, we are interested in expanding it to include other guidelines like AACE. We also implemented our model for the recommendation using SIO and that is still an ongoing process. We currently adopted DMTO’s style of using SWRL rules, but some rules can be replaced with restrictions on properties, and this is also something we would like to look into as future work. We would be very interested in having subject matter experts assist us in (a) extending our set of sample patients, (b) identifying the right course of action(s) that could help those patients, and (c) evaluating the recommendations that the system suggests for those patients to help
us improve on the results.
Cohort
We currently have two representations for the descriptive statistics on the cohorts ( patient populations ) utilized in research studies. One approach is rooted in classic OWL principles and uses the notion of associating every instance with a name and interconnecting statistics to the features via hasMeasure properties, however, this might not translate figuratively for a layman as we associate measures with characteristics like Disease. Our second approach is inspired by James McCusker’s paper on “A Provenance-Driven Semantics of Aggregation”, and we are instantiating StudyGroup’s as owl:Class and associating descriptions and restrictions on the same. For the second approach, you could help in automatically identifying features which might count as additional class membership restrictions like for a MalePatient set we only want to allow male patients. We would like feedback on which approach is more intuitive, and if neither what other approaches to represent aggregative values would be feasible. We would like some pointers to tools that are capable of extracting tables ( Table1’s containing population descriptions ) from PDFs ( of research studies) and do a good job at doing so.
Maintenance Policy
Team Maintenance:
- Any/all changes to the use case, concept map, ontology, and SPARQL queries will be made available on the Diabetes Treatment Support Ontology website
- Archives of the changes made to the use case, concept map, ontology, and SPARQL queries will be categorized and present on the website.
- The following hierarchy will be enforced for changes made to artifacts: use case --> concept map --> ontology --> SPARQL queries. If any changes are made to artifacts higher in the hierarchy (e.g., the use case is highest), changes should be cascaded to all artifacts lower in the hierarchy
Community Maintenance:
- Ontology permissions are as stated in the MIT License provide below (these allow for forking and branches of the ontology to be made for all permissible reasons)
- Members of the ontology team carry permission to make changes to the site; however, suggestions are welcome at: diabetestreatmentsupport@lists.rpi.edu
- Additional changes to the ontology are welcomed however to be accepted the ontology must successfully answer the competency questions present on the website (failure to do so indicates fundamental structure errors in the ontology that need to be submitted).
- The code should be accompanied by tests and documentation.
- One branch per feature or fix with sufficient commit messages.
- Ontology must have a version number, creator credits, and modification data present in the annotation header.
Project License:
MIT License
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
This was developed as a part of the Ontology Engineering course supervised by Prof. Deborah McGuinness and Ms. Elisa Kendall at RPI in Fall'18.